
|
||||||||||
|
||||||||||

Duplex-specific nuclease
SUPPORT |
|
||||||||||||||
|
Duplex-specific nuclease (DSN) is an enzyme purified from hepatopancreas of Red King (Kamchatka) crab (Shagin et al., 2002). DSN shows a strong preference for cleaving double-stranded (ds) DNA and DNA in DNA-RNA hybrid duplexes, and is practically inactive towards single-stranded (ss) DNA or single or double-stranded RNA. Moreover, the enzyme is capable to discriminate between perfectly and nonperfectly matched short DNA duplexes. DSN is widely used for full-length enriched cDNA normalization (Zhulidov et al., 2004; Bogdanova et al., 2011a) and for construction of normalized RNA-seq libraries for next generation sequencing ((Christodoulou et al., 2011), Illumina DSN normalization protocol). The list of approved DSN applications also includes normalization of genomic DNA (Shagina et al., 2010), cDNA depletion and ribosomal cDNA depletion (Bogdanova et al., 2009; Bogdanova et al., 2011b; Yi et al., 2011), cDNA subtraction (Peng et al., 2008), SNP detection (Shagin et al., 2002; Liu et al., 2011), construction of repeat-free FISH probes (Swennenhuis et al., 2012), multiplexed fluorescence detection of miRNAs (Yin et al., 2012), quantitative telomeric overhang determination (Zhao et al., 2008; Zhao et al., 2011), etc. |
Main properties of DSN
Substrate specificity: DSN exhibits strong cleavage preference for ds DNA substrates. It can show minor activity against ss DNA when both DSN enzyme and substrate are present at high concentrations, however this activity is not detectable in presence of competitive ds DNA and RNA (Zhao et al., 2008). DSN does not cleave RNA, but effectively cleaves DNA in DNA-RNA hybrid duplexes (Shagin et al., 2002)
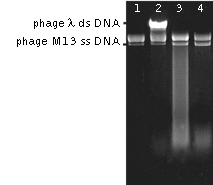 | Action of DSN on ss DNA of phage M13 and ds DNA of phage λ.Lanes 1, 2 – negative controls, incubation without nuclease. Lane 1: phage M13 DNA alone, Lane 2 – mixture containing phage M13 and λ DNA. Lanes 3; 4 – digestion of phage M13 and λ DNA mixture by DSN at 70°C for 1.5 min (lane 3) and 5 min (lane 4). |
|---|
Analysis of DSN activity on synthetic oligonucleotide substrates revealed that the enzyme discriminates between perfectly matched short DNA-DNA duplexes (10-12 bp) and duplexes of the same length with at least one mismatch. It requires at least 10 bp DNA or 15 bp DNA-RNA perfect duplex for cleavage.
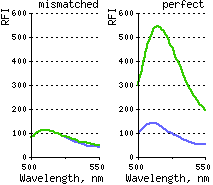 | DSN action on one mismatch-containing and perfectly matched DNA duplexes.Duplexes formed by 5-carboxyfluorescein (Fl)-5'-gccctatagt-3'-TAMRA signal probe and complementary targets (5’-actcactataCggcgaat-3’ and 5’-actcactatagggcgaat-3’) were incubated with DSN at 35°C for 15 min. Emission spectra were obtained on the spectrofluorimeter, with excitation at 480 nm. Dotted line – substrate fluorescence in the absence of enzyme; firm line – substrate fluorescence after incubation with DSN. |
|---|
Biochemical properties
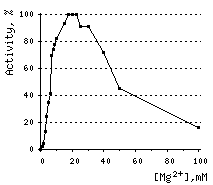
DSN acquires its enzymatic activity in the presence of divalent cations (Mn2+, Co2+, or Mg2+). Mg2+ ion concentration for most applications should be at least 5 mM. DSN is inhibited by EDTA.
The temperature optimum for DSN activity is 60°C. However, already at 80°C DSN retains only 10% of activity. This sharp decrease in activity may be attributable, at least in part, to ds DNA substrate denaturation.
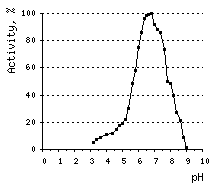
The optimal pH for DSN activity is 6.6. At pH values between 3 and 5, DSN displays only 10% of its maximal activity. The nuclease is stable at a wide range of pH (from 4 to 12) and temperatures below 65°C. About 60% of DSN activity remains after 30-min incubation at 70°C, and 40% – after incubation at 80°C.
 |  | 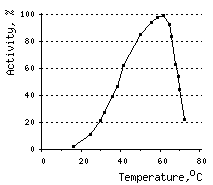 | |
|---|---|---|---|
Effects of various conditions on DSN activity.Activity of DNAse on ds DNA substrate was measured using modified Kunitz assay (Kunitz, 1950) | |||
Incubation of DSN with aggressive chemicals like 1% SDS, 10 mM β-mercaptoethanol, and 0.3% hydrogen peroxide at 37°C results in only a moderate drop in activity, after 30 min incubation about 90% of activity is preserved. However, a sharp decrease in activity was observed upon chemical treatment at 60°C. SDS completely inhibits DSN activity, while β-mercaptoethanol and hydrogen peroxide induce approximately 70% and 80% loss in activity, respectively.
DSN is highly sensitive to ionic force (e.g., a 10 times decrease in catalytic activity is observed in the presence of 0.2 M NaCl). The addition of chaotropic agents or polyamines to the reaction mixture also results in suppression of enzyme activity.
DSN is tolerant to proteinase K treatment (incubation at 37°C for 30 min).
References:
- Bogdanova EA, Barsova EV, Shagina IA, Scheglov A, Anisimova V, Vagner LL, Lukyanov SA, Shagin DA. Normalization of full-length-enriched cDNA. Methods Mol Biol. 2011a; 729 :85-98. doi: 10.1007/978-1-61779-065-2_6 / pmid: 21365485
- Bogdanova EA, Shagina IA, Mudrik E, Ivanov I, Amon P, Vagner LL, Lukyanov SA, Shagin DA. DSN depletion is a simple method to remove selected transcripts from cDNA populations. Mol Biotechnol. 2009; 41 ((3)):247-53. doi: 10.1007/s12033-008-9131-y / pmid: 19127453
- Bogdanova EA, Shagina IA, Yanushevich YG, Vagner LL, Lukyanov SA, Shagin DA. Preparation of prokaryotic cDNA for full-scale transcriptome analysis. Russian Journal of Bioorganic Chemistry. 2011b; 37 (6):775-8. ISI:000297344600012 http://www.springerlink.com
- Christodoulou DC, Gorham JM, Herman DS, Seidman JG. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr Protoc Mol Biol. 2011; Chapter 4 :Unit4.12. doi: 10.1002/0471142727.mb0412s94 / pmid: 21472699
- Kunitz M. Crystalline desoxyribonuclease; digestion of thymus nucleic acid; the kinetics of the reaction. J Gen Physiol. 1950; 33 :363-377. / pmid: 15406374
- Liu M, Yuan M, Lou X, Mao H, Zheng D, Zou R, Zou N, Tang X, Zhao J. Label-free optical detection of single-base mismatches by the combination of nuclease and gold nanoparticles. Biosens Bioelectron. 2011; 26 (11):4294-300. doi: 10.1016/j.bios.2011.04.014 / pmid: 21605966
- Peng RH, Xiong AS, Xue Y, Li X, Liu JG, Cai B, Yao QH. Kamchatka crab duplex-specific nuclease-mediated transcriptome subtraction method for identifying long cDNAs of differentially expressed genes. Anal Biochem. 2008; 372 (2):148-55. / pmid: 17905189
- Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002; 12 (12):1935-42. / pmid: 12466298
- Shagina I, Bogdanova E, Mamedov IZ, Lebedev Y, Lukyanov S, Shagin D. Normalization of genomic DNA using duplex-specific nuclease. Biotechniques. 2010; 48 (6):455-9. doi: 10.2144/000113422 / pmid: 20569220
- Swennenhuis JF, Foulk B, Coumans FA, Terstappen LW. Construction of repeat-free fluorescence in situ hybridization probes. Nucleic Acids Res. 2012; 40 (3):e20. doi: 10.1093/nar/gkr1123 / pmid: 22123742
- Yi H, Cho YJ, Won S, Lee JE, Jin Yu H, Kim S, Schroth GP, Luo S, Chun J. Duplex-specific nuclease efficiently removes rRNA for prokaryotic RNA-seq. Nucleic Acids Res. 2011; 39 (20):e140. doi: 10.1093/nar/gkr617 / pmid: 21880599
- Yin BC, Liu YQ, Ye BC. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J Am Chem Soc. 2012; 134 (11):5064-7. doi: 10.1021/ja300721s / pmid: 22394262
- Zhao Y, Hoshiyama H, Shay JW, Wright WE. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2008; 36 (3):e14. / pmid: 18073199
- Zhao Y, Shay JW, Wright WE. Telomere G-overhang length measurement method 1: the DSN method. Methods Mol Biol. 2011; 735 :47-54. doi: 10.1007/978-1-61779-092-8_5 / pmid: 21461810
- Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, Lukyanov SA, Shagin DA. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004; 32 (3):e37. / pmid: 14973331
|
Copyright 2002-2023 Evrogen. All rights reserved. Evrogen JSC, 16/10 Miklukho-Maklaya str., Moscow, Russia, Tel +7(495)988-4084, Fax +7(495)988-4085, e-mail:evrogen@evrogen.com |


