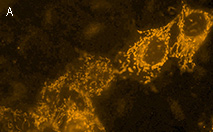
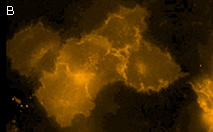
|
||||||||||
|
||||||||||

KillerOrange
SUPPORT |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
KillerOrange represents a mutant of KillerRed with a bright orange fluorescence (excitation maximum at
The blue-shifted spectrum of KillerOrange makes it potentially more suitable for two-photon microscopy than the parental KillerRed. Also, the large Stokes shift of over KillerOrange-KillerRed pair can potentially be used to independently ablate two cell populations. This pair also promises the orthogonal optical control of the propagation of signaling cascades either by chromophore-assisted light inactivation of the participating proteins or by triggering cascades with hydrogen peroxide produced by KillerRed and likely by KillerOrange. KillerOrange-KillerRed tandem fusions or combination of various photosensitizers in one cassette may enhance phototoxicity under white light irradiation and may be useful as a research tool in biology. |
Main properties
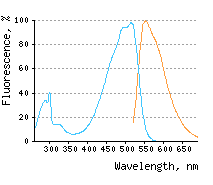
Blue line – excitation
|
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Recommended antibodies, filter sets, and activating parameters
KillerOrange can be recognized using Anti-KillerRed antibody (Cat.# AB961) available from Evrogen.
KillerOrange phototoxicity can be induced by blue or green light irradiation
Performance and use
KillerOrange can be used for in vivo killing of selected cells. It can be expressed and induced in various experimental systems, including bacteria and mammalian cells.
KillerOrange's suitability for light-induced killing of prokaryotic cells has been demonstrated using
Aliquots of
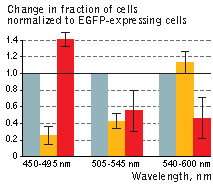 | KillerOrange (KO) and KillerRed (KR) toxicity under light illumination at different wavelengths.
Illumination with Grey column – EGFP, orange column – KillerOrange, red column – KillerRed. Error bars represent SD, N=4. |
|---|
KillerOrange-mediated killing of eukaryotic cells:
HEK 293 cells were grown in DMEM medium on 6-cm Petri dishes until about 70% confluency. They were then transiently transfected using FuGene HD (Roche) with one of the following plasmids: pKillerRed-dMito, pKillerOrange-dMito (Evrogen, Cat.# FP224) or pEGFP-N1 (Clontech).
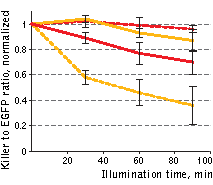 | Phototoxicity of mitochondria-targeted versions of KillerOrange and KillerRed in mammalian cells.
KillerOrange, KillerRed and EGFP-expressing cells were mixed and illuminated with Red dashed line – KillerRed, 447 nm; orange dashed line – KillerOrange, 447 nm; red line – KillerRed, 590 nm; orange line – KillerOrange, 590 nm. Error bars represent SD, N=3. |
|---|
A strong decrease in the fraction of KillerRed-expressing cells was seen in the population that passed through the orange light irradiation. In contrast, KillerOrange-expressing cells were substantially removed from the population of cells that was illuminated with the blue light. Taken together, these results show that KillerOrange is phototoxic to mammalian cells and that KillerRed and KillerOrange can be used simultaneously to independently control fates of two cell populations.
| Variant | Description | Related vector | Cat.# | ||
|---|---|---|---|---|---|
 | |||||
| Humanized KillerOrange |
KillerOrange is developed on the basis of the KillerRed [Bulina et al., 2006].
Its codon usage is optimized for high expression in mammalian cells [Haas et al.,
1996], but it can be successfully expressed in many other heterological systems. Mammalian expression vectors allowing fusions to the KillerRed C-terminus / N-terminus respectively |
|
FP221 | ||
|
|
FP222 | ||||
| KillerOrange-dMito fusion | Mitochondrial targeting sequence (MTS) was derived from subunit VIII precursor of human cytochrome C oxidase [Rizzuto et al., 1989; Rizzuto et al., 1995]. Two MTS were fused to KillerOrange N-terminus. When expressed in mammalian cells, this variant is localized in mitochondria. |
|
FP224 | ||
References:
- Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006; 24 (1):95-9. / pmid: 16369538
- Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996; 6 (3):315-24. / pmid: 8805248
- Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995; 5 (6):635-42. / pmid: 7552174
- Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi GM, Mengel T, Walsh F, Kadenbach B, DiMauro S, Schon EA. A gene specifying subunit VIII of human cytochrome c oxidase is localized to chromosome 11 and is expressed in both muscle and non-muscle tissues. J Biol Chem. 1989; 264 (18):10595-600. / pmid: 2543673
- Sarkisyan KS, Zlobovskaya OA, Gorbachev DA, Bozhanova NG, Sharonov GV, Staroverov DB, Egorov ES, Ryabova AV, Solntsev KM, Mishin AS, Lukyanov KA. KillerOrange, a Genetically Encoded Photosensitizer Activated by Blue and Green Light. PLoS One. 2015; 10 (12):e0145287. doi: 10.1371/journal.pone.0145287 / pmid: 26679300
- Skene JH, Virag I. Posttranslational membrane attachment and dynamic fatty acylation of a neuronal growth cone protein, GAP-43. J Cell Biol. 1989; 108 (2):613-24. / pmid: 2918027
|
Copyright 2002-2023 Evrogen. All rights reserved. Evrogen JSC, 16/10 Miklukho-Maklaya str., Moscow, Russia, Tel +7(495)988-4084, Fax +7(495)988-4085, e-mail:evrogen@evrogen.com |