|
||||||||||
|
||||||||||

KillerRed
SUPPORT |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Photosensitizers are chromophores that generate reactive oxygen species (ROS) upon light irradiation. They can be used for precise inactivation of selected proteins in chromophore-assisted light inactivation (CALI) technique and for the light-induced cell killing, for example in photodynamic therapy. Besides KillerRed protein (and its derivative KillerOrange), all known to date photosensitizers require chemical compounds that should be introduced into living systems exogenously. Red fluorescent protein KillerRed is the first genetically-encoded photosensitizer [Bulina et al., 2006a]. Unlike chemical analogs, KillerRed can be directly expressed by target cells, both individually and in fusion with a target protein. It shows no cell toxic effects before light activation. Upon green or orange light irradiation, KillerRed generates ROS that damage the neighboring molecules. |
Main properties
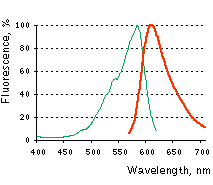
Normalized excitation (green line) and emission (red line) spectra. |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Recommended antibodies, filter sets, and activating parameters
KillerRed can be recognized using Anti-KillerRed antibody (Cat.# AB961) available from Evrogen.
Before light activation, KillerRed can be detected using TRITC filter set or similar. Recommended Omega Optical filter sets are QMAX-Red and XF174.
KillerRed phototoxicity is induced by green or orange light irradiation at 540-590 nm and depends on light intensity irradiation time and KillerRed concentration and localization. Arc-lamp irradiation or LEDs light is strongly recommended; laser-light irradiation in confocal mode is much less efficient.
In CALI, mild illumination of KillerRed-tagged protein for a limited time results in precise inactivation of this protein only. Upon more prolonged and intensive irradiation, KillerRed can be effectively used for damaging the organelles and killing the target cells. Light intensity and irradiation time should be individually determined for particular biological system and instrumentation.
KillerRed-mediated ROS production is accompanied by profound KillerRed photobleaching. The resulting cell events (cell fate after irradiation, effect on protein localization) can be monitored using another fluorescent reporter, for example a green fluorescent protein. We recommend that you use TurboGFP for cell and organelle, or TagGFP2 for protein labeling.
Performance and use
KillerRed can be used for in vivo killing of selected cells and CALI applications. It can be expressed and induced in various experimental systems, including bacteria, Xenopus, zebrafish, and mammalian cells.
KillerRed is 2000-fold more phototoxic than EGFP; however, it is not as effective as chemical probes. KillerRed-mediated ROS production is accompanied by profound KillerRed photobleaching.
Despite it dimerization capacity, KillerRed demonstrates successful performance in many fusions including that with mitochondrial targeted signal, cytoplasmic
KillerRed suitability to generate stably transfected cells has been proven by Marinpharm company.
KillerRed's suitability for light-induced killing of prokaryotic cells has been demonstrated using
KillerRed killed 96% of bacterial cells after 10 min and almost 100% of cells after 20 min of irradiation with white light.
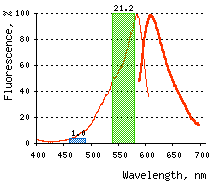 | 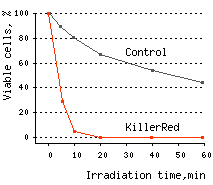 | KillerRed-mediated killing of bacteria cells upon irradiation with different wavelength.
Left graph: Blue and green rectangles show relative phototoxic effect from irradiation with blue (460-490 nm) and green (540-580 nm) light of 35 mW/cm2. Numbers above the rectangles represent decrease in viable
Right graph: Time-course of white light-induced (1 W/cm2) killing of |
|---|
KillerRed-mediated killing of eukaryotic cells: The following two ways have been found to be effective for killing the eukaryotic cells using KillerRed: (1) via an apoptotic pathway using KillerRed targeted to mitochondria, and (2) via membrane lipid oxidation using membrane-localized KillerRed. Irradiation of KillerRed localized in cell cytosol has a weak effect on cell survival.
Effects of KillerRed localized in mitochondria: Mitochondrial localization increases the phototoxic effect of photosensitizers (primarily by provoking the apoptosis). Use of KillerRed targeted to mitochondria allows effective cell killing through an apoptotic pathway as has been demonstrated in the following experiments:
HeLa cells expressing cytoplasmic TurboGFP and mitochondria-localized KillerRed-dMito were generated. 10-min (515-560 nm excitation filter, 7 W/cm2) irradiation of selected cells with green light resulted in profound KillerRed photobleaching. 60 min after irradiation, cells had an abnormal shape and "bubbles" typical of apoptotic pathway. These cells disrupted within the next 30-60 min.
In another experiment, nearly 100% of B16 melanoma cells expressing KillerRed targeted to mitochondria died within 45 min after 15-min of irradiation (40x objective, 535-575 nm excitation filter, 3.3 W/cm2). It is noticeable that when preincubated with the pancaspase inhibitor zVAD-fmk (10 μM), the cells were resistant to the same light exposure and held their shape for at least 1.5 hours after illumination.
Apart from the immediate phototoxic effect, photosensitizers can mediate postponed cellular responses such as cell growth arrest or cell death via long-term apoptotic mechanism:
B16 melanoma cells expressing mitochondria-targeted KillerRed or EGFP were mixed together and irradiated by green light of low intensity (3.7x objective, 535-575 nm excitation filter, 115 mW/cm2) for 45 min [Bulina et al., 2006a]. No red fluorescent cells were observed in 16 hrs after irradiation, whereas green fluorescent cells remained viable. It confirms that mitochondria-localized KillerRed can mediate cell death through long-term mechanisms in response to low-intensity illumination. This effect can be used in different applications.
Effects of KillerRed localized in membrane: Comparing with the mitochondria-targeted KillerRed, irradiation of membrane-localized KillerRed causes a more effective and fast cell death within 10-30 min, presumably because of lipid oxidation [Bulina et al., 2006b].
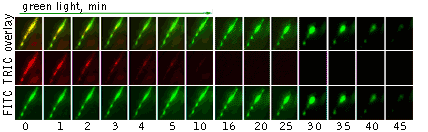
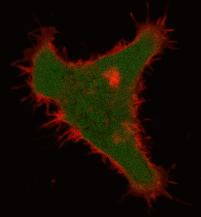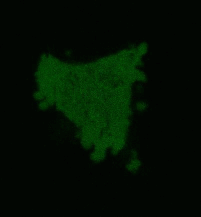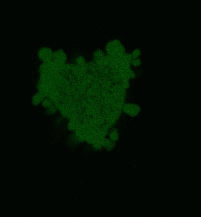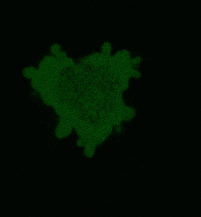 | Light-induced killing of HeLa cell using membrane-targeted KillerRed (movie).First image: intact cell features red fluorescent cellular membrane and green fluorescent cytosol due to membrane-targeted KillerRed and TurboGFP expression, respectively. Further images: 40 minute time-course of cell fragmentation induced by 10-min green light (laser 543 nm, 300 mW/cm2) irradiation. Movie from Bulina et al., 2006b. |
|---|
Moreover, membrane-targeted KillerRed was shown to be an effective tool for the light-induced cell killing within a developing zebrafish. Zebrafish embryo was microinjected with a mixture of vectors driving expression of membrane-targeted KillerRed and a green fluorescent protein at the single- cell stage. A muscle cell expressing both proteins was irradiated with green light (40x objective, TRITC filter set, 10 min) at 48 hrs after fertilization. By the end of 10-min irradiation, the cell already started to change its shape. Within 20 min after irradiation was stopped, the cell was disrupted completely. Mitochondria targeted KillerRed was shown to be of low efficiency in similar experiments.
 |  | |
|---|---|---|
Light-induced killing of a muscle cell within a developing zebrafish embryo.Left image: A region expressing membrane-targeted KillerRed and green fluorescent marker; right image: time course of light-induced killing of a muscle cell within a developing zebrafish. Fluorescence was collected using standard FITC and TRITC filter sets. | ||
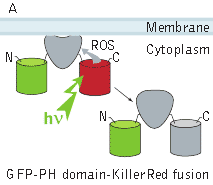
KillerRed use for CALI applications: KillerRed use for CALI application has been demonstrated on the model of β-galactosidase inactivation in bacterial cells and inactivation of pleckstrin homology (PH) domain of phospholipase C delta-1 (PLC delta-1) in mammalian cells. In the first experiment, KillerRed was fused to β-galactosidase (β-gal) enzyme and expressed in
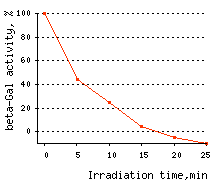 | Time-course of CALI of β-galactosidase.
|
|---|
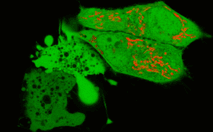
In the second experiment, a triple EGFP-PH-KillerRed fusion protein that allows both protein visualization and CALI was transiently expressed in mammalian cell line. Intracellular localization of EGFP signal was evaluated before and after CALI of the PH domain using confocal and fluorescence microscopy. In intact cells, the fusion is located predominantly at the plasma membrane because of specific affinity of PH domain to phosphatidylinositol 4,5-bisphosphate.
Irradiation with intense green light led to KillerRed-mediated ROS production, PH domain damage, and fusion protein dissociation from the membrane. After 10 sec of green light irradiation (63x objective, mercury lamp, 515-560 nm filter, 7W/cm2), translocation of the PH domain into cytosol was clearly visible. When irradiated for a longer period of time, considerable amount of PH domain translocated into cytosol, increasing the cytoplasm-to-membrane green fluorescent signal ratio to 0.5-0.9.
In the negative control experiments, the cellular location of a DsRed-Express (Clontech) containing construct, GFP-PH-DsRed-Express, showed no dependence on green light irradiation. Similarly, no detectable CALI of the PH domain was achieved when KillerRed was expressed in the cell separately from PH domain, in either the membrane or cytosol.
| Variant | Description | Related vector | Cat.# | |
|---|---|---|---|---|
 | ||||
| Humanized KillerRed | KillerRed id developed on the basis of the chromoprotein from Anthomedusae sp.[Shagin et al., 2004]. Its codon usage is optimized for high expression in mammalian cells [Haas et al., 1996], but it can be successfully expressed in many other heterological systems. |
|
FP962 | |
|
|
FP963 | |||
| KillerRed-mem fusion | KillerRed is fused with membrane localization signal (MLS) of neuromodulin. The neuromodulin MLS (N-terminal 20 amino acid residues) contains a signal for posttranslational palmitoylation of cysteines 3 and 4 that targets KillerRed to cellular membranes [Skene and Virag, 1989]. Irradiation of membrane-localized KillerRed leads to high effective and fast cell death, presumably due to lipid oxidation. Comparing to the mitochondrially targeted KillerRed, irradiation of membrane-localized KillerRed leads to even more effective and fast cell death (within 10-30 min). Membrane-targeted KillerRed was shown to be suitable for the light induced cell killing within a developing zebrafish. |
|
FP966 | |
References:
- Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006a; 24 (1):95-9. / pmid: 16369538
- Bulina ME, Lukyanov KA, Britanova OV, Onichtchouk D, Lukyanov S, Chudakov DM. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat Protoc. 2006b; 1 (2):947-53. / pmid: 17406328
- Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996; 6 (3):315-24. / pmid: 8805248
- Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995; 5 (6):635-42. / pmid: 7552174
- Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi GM, Mengel T, Walsh F, Kadenbach B, DiMauro S, Schon EA. A gene specifying subunit VIII of human cytochrome c oxidase is localized to chromosome 11 and is expressed in both muscle and non-muscle tissues. J Biol Chem. 1989; 264 (18):10595-600. / pmid: 2543673
- Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, Semenova TN, Ugalde JA, Meyers A, Nunez JM, Widder EA, Lukyanov SA, Matz MV. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol. 2004; 21 (5):841-50. / pmid: 14963095
- Skene JH, Virag I. Posttranslational membrane attachment and dynamic fatty acylation of a neuronal growth cone protein, GAP-43. J Cell Biol. 1989; 108 (2):613-24. / pmid: 2918027
|
Copyright 2002-2023 Evrogen. All rights reserved. Evrogen JSC, 16/10 Miklukho-Maklaya str., Moscow, Russia, Tel +7(495)988-4084, Fax +7(495)988-4085, e-mail:evrogen@evrogen.com |